Publications
Discover recent publications from our research on winter cereals, advancing knowledge in crop genetics, physiology, and adaptation.
Lab Publications

Comparative study of winter cereals forage productivity in Mediterranean agro-system highlight possibilities for crop improvement and agronomic diversification

Stem traits promote wheat climate-resilience

New flavors from old wheats: exploring the aroma profiles and sensory attributes of local Mediterranean wheat landraces
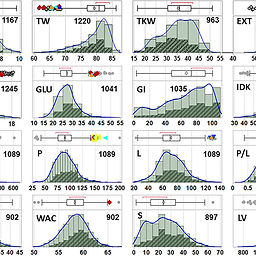
Towards stable wheat grain yield and quality under climatic instability
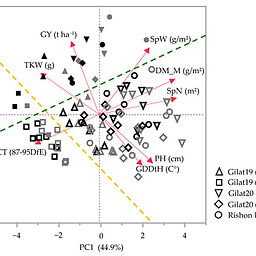
In-Field Comparative Study of Landraces vs. Modern Wheat Genotypes under a Mediterranean Climate
Collaborative Publications

UAV-borne hyperspectral and thermal imagery integration empowers genetic dissection of wheat stomatal conductance
April 19, 2025

The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat
May 23, 2019

Whole-Genome Sequencing of the Wild Barley Diversity Collection: A Resource for Identifying and Exploiting Genetic Variation for Cultivated Barley Improvement
November 20, 2024

Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69
January 8, 2024

Genome Based Meta-QTL Analysis of Grain Weight in Tetraploid Wheat Identifies Rare Alleles of GRF4 Associated with Larger Grains
December 17, 2018